Sitemap
A list of all the posts and pages found on the site. For you robots out there is an XML version available for digesting as well.
Pages
Posts
Future Blog Post
Published:
This post will show up by default. To disable scheduling of future posts, edit config.yml and set future: false.
Blog Post number 4
Published:
This is a sample blog post. Lorem ipsum I can’t remember the rest of lorem ipsum and don’t have an internet connection right now. Testing testing testing this blog post. Blog posts are cool.
Blog Post number 3
Published:
This is a sample blog post. Lorem ipsum I can’t remember the rest of lorem ipsum and don’t have an internet connection right now. Testing testing testing this blog post. Blog posts are cool.
Blog Post number 2
Published:
This is a sample blog post. Lorem ipsum I can’t remember the rest of lorem ipsum and don’t have an internet connection right now. Testing testing testing this blog post. Blog posts are cool.
Blog Post number 1
Published:
This is a sample blog post. Lorem ipsum I can’t remember the rest of lorem ipsum and don’t have an internet connection right now. Testing testing testing this blog post. Blog posts are cool.
publications
Multi-omics Integration based Predictive Model for Survival Prediction of Lung Adenocarcinoma
Published in 2019 Grace Hopper Celebration India (GHCI), 2019
This paper presents a multi-omics integration model for survival prediction in lung adenocarcinoma.
Recommended citation: Malik, Vidhi, Dutta, Shayoni, Kalakoti, Yogesh, others. (2019). "Multi-omics Integration based Predictive Model for Survival Prediction of Lung Adenocarcinoma." 2019 Grace Hopper Celebration India (GHCI).
Deep learning assisted multi-omics integration for survival and drug-response prediction in breast cancer
Published in BMC genomics, 2021
This paper explores deep learning for integrating multi-omics data to predict survival and drug response in breast cancer.
Recommended citation: Malik, Vidhi, Kalakoti, Yogesh, Sundar, Durai. (2021). "Deep learning assisted multi-omics integration for survival and drug-response prediction in breast cancer." BMC genomics. 22(1), 214. https://doi.org/10.1186/s12864-021-07524-2
SurvCNN: a discrete time-to-event cancer survival estimation framework using image representations of omics data
Published in Cancers, 2021
This paper presents SurvCNN, a framework for cancer survival estimation using image representations of omics data.
Recommended citation: Kalakoti, Yogesh, Yadav, Shashank, Sundar, Durai. (2021). "SurvCNN: a discrete time-to-event cancer survival estimation framework using image representations of omics data." Cancers. 13(13), 3106.
SurvCNN: a discrete time-to-event cancer survival estimation framework using image representations of omics data
Published in Cancers, 2021
This paper presents SurvCNN, a framework for cancer survival estimation using image representations of omics data.
Recommended citation: Kalakoti, Yogesh, Yadav, Shashank, Sundar, Durai. (2021). "SurvCNN: a discrete time-to-event cancer survival estimation framework using image representations of omics data." Cancers. 13(13), 3106.
TransDTI: transformer-based language models for estimating DTIs and building a drug recommendation workflow
Published in ACS omega, 2022
This paper discusses TransDTI, using transformer-based models for drug-target interaction estimation and drug recommendation.
Recommended citation: Kalakoti, Yogesh, Yadav, Shashank, Sundar, Durai. (2022). "TransDTI: transformer-based language models for estimating DTIs and building a drug recommendation workflow." ACS omega. 7(3), 2706-2717.
TransDTI: transformer-based language models for estimating DTIs and building a drug recommendation workflow
Published in ACS omega, 2022
This paper discusses TransDTI, using transformer-based models for drug-target interaction estimation and drug recommendation.
Recommended citation: Kalakoti, Yogesh, Yadav, Shashank, Sundar, Durai. (2022). "TransDTI: transformer-based language models for estimating DTIs and building a drug recommendation workflow." ACS omega. 7(3), 2706-2717.
Deep neural network-assisted drug recommendation systems for identifying potential drug–target interactions
Published in ACS omega, 2022
This paper investigates deep neural networks for drug recommendation by identifying drug-target interactions.
Recommended citation: Kalakoti, Yogesh, Yadav, Shashank, Sundar, Durai. (2022). "Deep neural network-assisted drug recommendation systems for identifying potential drug--target interactions." ACS omega. 7(14), 12138-12146.
Genome Sequence of a Potent Biosurfactant-Producing Bacterium, Franconibacter sp. Strain IITDAS19
Published in Microbiology Resource Announcements, 2022
This paper reports the genome sequence of Franconibacter sp. IITDAS19, a biosurfactant-producing bacterium.
Recommended citation: Sharma, Jyoti, Kalakoti, Yogesh, Srivastava, Preeti, Sundar, Durai. (2022). "Genome Sequence of a Potent Biosurfactant-Producing Bacterium, Franconibacter sp. Strain IITDAS19." Microbiology Resource Announcements. 11(8), e00419-22.
Computational Methods and Deep Learning for Elucidating Protein Interaction Networks
Published in Computational Biology and Machine Learning for Metabolic Engineering and Synthetic Biology, 2022
This chapter explores computational methods and deep learning for analyzing protein interaction networks.
Recommended citation: Vora, Dhvani Sandip, Kalakoti, Yogesh, Sundar, Durai. (2022). "Computational Methods and Deep Learning for Elucidating Protein Interaction Networks." Computational Biology and Machine Learning for Metabolic Engineering and Synthetic Biology. 285-323.
Modulation of DNA-protein Interactions by Proximal Genetic Elements as Uncovered by Interpretable Deep Learning
Published in Journal of Molecular Biology, 2023
This paper examines DNA-protein interactions modulated by genetic elements using interpretable deep learning.
Recommended citation: Kalakoti, Yogesh, Peter, Swathik Clarancia, Gawande, Swaraj, Sundar, Durai. (2023). "Modulation of DNA-protein Interactions by Proximal Genetic Elements as Uncovered by Interpretable Deep Learning." Journal of Molecular Biology. 435(13), 168121.
M-Ionic: prediction of metal-ion-binding sites from sequence using residue embeddings
Published in Bioinformatics, 2024
This paper discusses predicting metal-ion-binding sites from sequence using residue embeddings.
Recommended citation: Shenoy, Aditi, Kalakoti, Yogesh, Sundar, Durai, Elofsson, Arne. (2024). "M-Ionic: prediction of metal-ion-binding sites from sequence using residue embeddings." Bioinformatics. 40(1), btad782. http://dx.doi.org/10.1093/bioinformatics/btad782
Estimating protein–ligand interactions with geometric deep learning and mixture density models
Published in Journal of Biosciences, 2024
This paper focuses on estimating protein-ligand interactions using geometric deep learning and mixture density models.
Recommended citation: Eliot, Kalakoti, Yogesh, Gawande, Swaraj, Sundar, Durai. (2024). "Estimating protein--ligand interactions with geometric deep learning and mixture density models." Journal of Biosciences. 49(4), 97.
Prediction of structural variation
Published in Current opinion in structural biology, 2025
This paper focuses on predicting structural variations in biological systems.
Recommended citation: Kalakoti, Yogesh, Sanjeev, Airy, Wallner, Björn. (2025). "Prediction of structural variation." Current opinion in structural biology. 91, 103003.
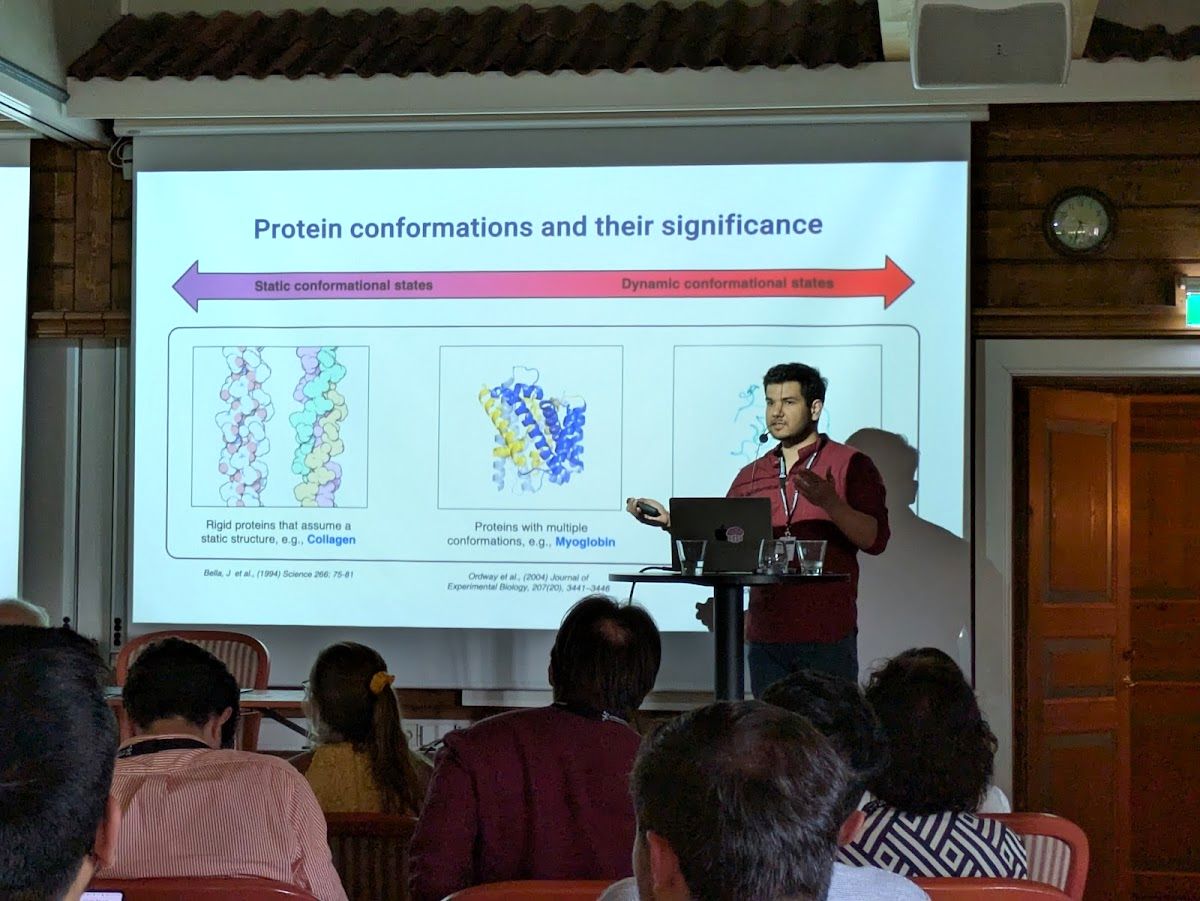
AFsample2 predicts multiple conformations and ensembles with AlphaFold2
Published in Communications Biology, 2025
This paper discusses AFsample2, a tool for predicting multiple conformations using AlphaFold2.
Recommended citation: Kalakoti, Yogesh, Wallner, Björn. (2025). "AFsample2 predicts multiple conformations and ensembles with AlphaFold2." Communications Biology. 8(1), 373.
research
Postdoctoral research
Linköping University, Sweden 
Portfolio item number 2
Short description of portfolio item number 2 
Post-Doctoral research
Indian Institure of Technology Delhi, Delhi 
talks
teaching
Teaching experience 1
Undergraduate course, University 1, Department, 2014
This is a description of a teaching experience. You can use markdown like any other post.
Teaching experience 2
Workshop, University 1, Department, 2015
This is a description of a teaching experience. You can use markdown like any other post.